In the first part, we’ve introduced the fundamental ideas on MLP and its training method Backpropagation. In this part, let’s get started to implement those theories to come up with a keras-style MLP.
We begin with a standard import:
import numpy as np
import matplotlib.pyplot as plt
First of all, we define some activation functions and their derivatives:
def sigmoid(Z):
return 1 / (1 + np.exp(-np.clip(Z, -250, 250)))
def relu(Z):
return np.maximum(0, Z)
def sigmoid_bk(Z):
sig = sigmoid(Z)
return sig * (1 - sig)
def relu_bk(Z):
return np.where(Z <=0, 0, 1)
act_funcs = {'sigmoid':sigmoid, 'relu':relu}
back_act = {'sigmoid': sigmoid_bk, 'relu': relu_bk}
For each layer, we need to remember the number of units, what’s the activation function, weights $W$, bias $b$, the linear combination $z$, and the output from activation function $a$. So we create a module enabling those methods:
class Layer(object):
def __init__(self, units, activation):
self.units = units
self.activation = act_funcs[activation]
self.back_act = back_act[activation]
self.a_h = None
self.z_h = None
self.W = None
self.b = None
Module Model contains the architecture of the model as well as the training processes:
class Model(object):
def __init__(self, lr, epochs, batch_size=200, seed=123):
self.arch = []
self.lr = lr
self.epochs = epochs
self.rnd = np.random.RandomState(seed)
self.batch_size = batch_size
self.cost = []
self.fit_flag = False
def init_w(self):
n_layers = len(self.arch)
for i in range(1, n_layers):
layer0 = self.arch[i-1]
layer1 = self.arch[i]
w_ = self.rnd.randn(layer0.units, layer1.units)
b_ = np.zeros(layer1.units)
layer1.W = w_
layer1.b = b_
def init_az(self):
for layer in self.arch:
layer.a_h = None
layer.z_h = None
def add(self, layer):
self.arch.append(layer)
def _compute_cost(self, y, output):
term1 = -np.dot(y.T, np.log(output))
term2 = np.dot((1- y).T, np.log(1 - output))
cost = (term1 + term2)[0,0]
dA_init = -(np.divide(y, output) - np.divide(1 - y, 1 - output))
return cost, dA_init
def _forward(self, layer0, layer1):
a_h0 = layer0.a_h
z_h1 = np.dot(a_h0, layer1.W) + layer1.b
a_h1 = layer1.activation(z_h1)
layer1.z_h = z_h1
layer1.a_h = a_h1
def _backward(self, layer0, layer1, dA):
dW = np.dot(layer0.a_h.T, layer1.back_act(layer1.z_h) * dA)
db = np.sum(layer1.back_act(layer1.z_h) * dA, axis=0, keepdims=True)
dA = np.dot(layer1.back_act(layer1.z_h) * dA, layer1.W.T)
layer1.W = layer1.W - self.lr * dW
layer1.b = layer1.b - self.lr * db
return dA
def fit(self, X, y, retrain=True):
#########################
#init all Ws and set a_h0 = X
#########################
idx = [i for i in range(len(X))]
self.rnd.shuffle(idx)
idx = idx[:self.batch_size]
n_layers = len(self.arch)
self.init_w()
if retrain:
self.init_az()
if X.shape[1] == self.arch[0].units:
self.arch[0].a_h = X[idx]
else:
raise Exception("Input shape is not right")
for e in range(self.epochs):
#######################
# forward propra
#######################
for i in range(1, n_layers):
layer0 = self.arch[i - 1]
layer1 = self.arch[i]
self._forward(layer0, layer1)
cost, dA_init = self._compute_cost(y[idx], self.arch[-1].a_h)
self.cost.append(cost)
dA = dA_init
#######################
# backward propra
#######################
for j in range(1, n_layers):
layer0 = self.arch[-j - 1]
layer1 = self.arch[-j]
dA = self._backward(layer0, layer1, dA)
self.fit_flag = True
return self
def predict(self, X):
if self.fit_flag:
n_layers = len(self.arch)
for i in range(1, n_layers):
layer = self.arch[i]
Z = np.dot(X, layer.W) + layer.b
X = layer.activation(Z)
self.fit_flag = True
return X
else:
print('The model has not been trained yet')
Couple of comments on the code:
- Since the model is designed for a binary classification model. We have used Log loss or called Cross-Entropy loss. Its expression is:
$-(ylog(haty)+(1-y)log(1-haty))$ (2.1)
where $haty$ is the output of our model. In essence, the equation (2.1) is the loss function we use to calculate costs in the function _compute_cost. We also compute $(delC_0)/(delA^((L)))$ or dA_init in the code. This value is calculated from the derivatives of our loss function (equation 2.1):
$-((y)/(haty) - (1-y)/(1-haty))$ (2.2) - We used SGD to train the model. Recall it from previous part, weights will be updated with a subset of the inputs. batch_size is used to control this size.
Try our model
In order to test if our model is working, we will use a MLP with hidden-layers of 4 to solve a binary classification problem from sklearn:
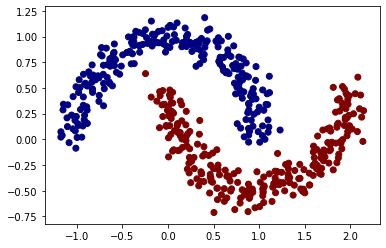
from sklearn.datasets import make_moons
X, y = make_moons(n_samples=500, noise=0.1)
y = y.reshape(-1, 1)
model_nn = Model(lr=0.0001, epochs=1000)
model_nn.add(Layer(2, 'relu'))
model_nn.add(Layer(4, 'relu'))
model_nn.add(Layer(8, 'relu'))
model_nn.add(Layer(8, 'relu'))
model_nn.add(Layer(5, 'relu'))
model_nn.add(Layer(1, 'sigmoid'))
model_nn.fit(X, y)
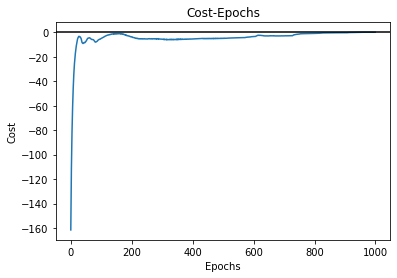
cost = model_nn.cost
plt.plot(range(len(cost)), cost)
plt.axhline(color='k')
plt.title("Cost-Epochs")
plt.xlabel('Epochs')
plt.ylabel('Cost')
We can also see how the cost changes as epoch increases:
Visualization the result by drawing decision boundary. In comparison, we also plot the result from MLP in sklearn with the same dataset:
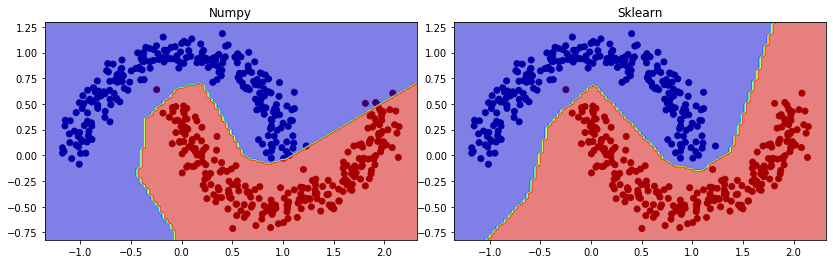
Not a bad job.
You can also review the code at
jupyter notebook code
